24 Working with Nodal Attributes in Network Models
Our examples so far were all built on a set of nodes that were homogeneous, except for infectious status. In this tutorial, we’ll introduce two groups that can systematically differ in their behavior and/or their epidemic parameters (e.g. average duration of infection). We’ll motivate our example with a heterosexual network in which the groups represent females and males in the population.
Download the R script to follow along with this tutorial here.
24.1 Getting Started
To get started, load the EpiModel library.
We also recommend that you clear your environment to avoid any leftover objects from the last tutorials or labs.
And we will set the seed to ensure reproducible results:
24.2 Network Model
For the network model parameterization, we’ll fit a two-group model with a differential degree distribution by group. We will do so using the special group attribute.
In core EpiModel, the group attribute has a special role for these built-in models that we are exploring in this course. It allows for easy parameterization of heterogeneous epidemic parameters (e.g., group-specific recovery rates). This functionality requires that group takes only two values, which must be 1 and 2. In our example, group 1 will represent women and group 2 men.
The group attribute makes life easier in this specific circumstance. In future examples, we can, and will, use other nodal attributes that can take more values (e.g. age or race) and model their impacts on network structure specifically. (Epidemic parameters can also be stratified by an arbitrary number of attributes with arbitrary values; however, this requires some additional programming of the epidemic modules in EpiModel. This is covered in the NME II workshop.)
24.2.1 Initialization
First we set up the network. The number of nodes in each group will be 250:
Network attributes:
vertices = 500
directed = FALSE
hyper = FALSE
loops = FALSE
multiple = FALSE
bipartite = FALSE
total edges= 0
missing edges= 0
non-missing edges= 0
Vertex attribute names:
vertex.names
No edge attributesWe next define our nodal attribute group, and then set it on the network. We can print out the network again to see that it has been added to the list of vertex attributes. Remember, to access the special functionality associated with the group attribute, its values must be 1 and 2 only.
Code
Network attributes:
vertices = 500
directed = FALSE
hyper = FALSE
loops = FALSE
multiple = FALSE
bipartite = FALSE
total edges= 0
missing edges= 0
non-missing edges= 0
Vertex attribute names:
group vertex.names
No edge attributesWe can use get_vertex_attribute to extract the same attribute from the network object:
[1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[112] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[149] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[186] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[223] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2
[260] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[297] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[334] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[371] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[408] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[445] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
[482] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 224.2.2 Model specification
We are going to specify:
- a constraint that all ties must be cross-sex
- the expected total number of ties
- the expected proportion of each sex that has concurrent ties at any point in time.
To do so, we begin by thinking through some basic logic to identify the rules that our model and its target statistics must obey.
First off, the total number of ties that females have with males must logically equal the total number of ties that males have with females. This logical rule is always the case when considering undirected ties between two groups.
The constraint above about all ties being cross-sex means that this logical relationship can be extended here to say that for this network, total number of ties for females must equal the the total number of ties for males. Note that this is not universally true, but only holds in this case because of the pure cross-sex tie constraint.
The total number of ties for a group equals the number of nodes in the group times the mean degree for that group. We specified above that the number of females and the number of males are equal. Therefore, we can further extend our logical rules to say that, for this network, the mean degree for females must equal the mean degree for males. Note again that this is not universally true, but only holds in this case because of both the pure cross-sex tie constraint and the fact that the two groups have the exact same population size.
We will thus set a single overall mean degree. We will chose a value of 0.66.
Now we would like to consider the proportion of each group that has concurrent ties at any point in time. Is there a logical constraint on how these numbers relate to each other? Think about it before proceeding….
In general, there is no simple constraint here. For example, think of a population of 4 women and 4 men. Each woman has one partner, and for all four of them it’s man #1. The women have 0% concurrency while the men have 25%. What if the first two women are partners with man #1 and the other two with man #2? Now the men have 50% concurrency while the women still have 0%. There can be overall constraints on concurrency imposed by the mean degree (e.g. if it’s greater than 1 then someone has to have concurrent partners). And there are constraints at the extremes on the sex-specific values. But in general, they are pretty free to move independently.
So, we will specify the proportion of each sex that has concurrent partners: 5% for females, and 15% for males. It is common in empirical data to see the value higher for males than females.
As you progress into more complex network models, it is helpful to practice thinking through the logical relationships among your terms and their associated statistics carefully. This helps to ensure that you are including the right set of terms for your goal, and not over- or under-specifying the model. We dive into this more deeply in NME-II.
24.2.3 Formation model
Next, we specify the terms in our formation model. As usual, we want an edges term to model the overall mean degree.
The nodematch term is new: it allows one to model assortative or disassortative mixing across an attribute. We will use it to encode our assumption that all ties are cross-sex.
The concurrent term specifies the proportion of nodes expected to have concurrent partners at any point in time. It includes an option by argument, which takes the name of a nodal attribute on which the effect can be disaggregated. To tell it that we want to specify each group, we need to set the levels argument. There are multiple equivalent ways we could do this, including passing the specific values 1:2. However, using NULL is a more general way to say “all levels” without having to know the number of their values:
Putting this all together, we get:
For the target statistics, we specify the number of edges as a function of mean degree. The target statistic for nodematch with a purely disassortative mixing model is 0: there are no edges between persons matched on the value of the group variable. Finally, we calculate the expected number of females and males with concurrent partners, respectively:
[1] 165.0 0.0 12.5 37.5Note that it is perfectly fine for our target values for concurrent to not be whole numbers. How can that be, when the number of people with concurrent partners must be a whole number at any point in time? Because the average of a set of whole numbers (in our case, averaged over time) does not need to be a whole number.
24.2.4 Dissolution model
The dissolution model is parameterized the same as the prior example but with a shorter partnership duration than the first tutorial; we’ll go with 20 steps:
24.2.5 Estimation
The netest function is used for network model fitting.
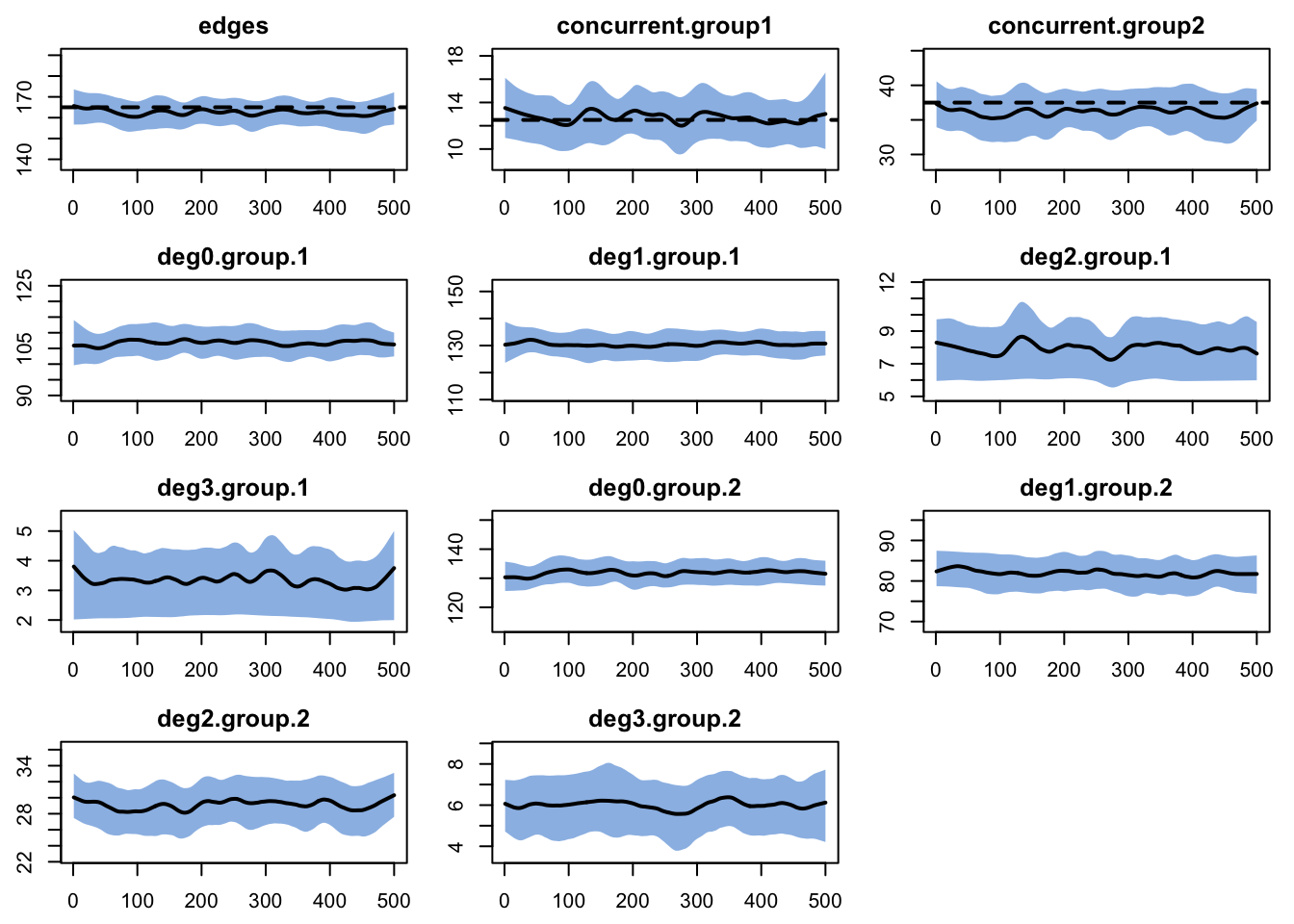
For the dynamic diagnostics, we simulate from the model fit. We’ll track not only the concurrent statistics but also the degree distribution as a whole:
Code
Network Diagnostics
-----------------------
- Simulating 25 networks
- Calculating formation statisticsEpiModel Network Diagnostics
=======================
Diagnostic Method: Dynamic
Simulations: 25
Time Steps per Sim: 500
Formation Diagnostics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means)
edges 165.0 162.557 -1.481 0.494 -4.945 3.060
concurrent.group1 12.5 12.711 1.690 0.161 1.309 1.090
concurrent.group2 37.5 36.166 -3.556 0.181 -7.369 1.123
deg0.group.1 NA 106.825 NA 0.283 NA 1.829
deg1.group.1 NA 130.463 NA 0.269 NA 1.750
deg2.group.1 NA 7.934 NA 0.087 NA 0.634
deg3.group.1 NA 3.323 NA 0.055 NA 0.384
deg0.group.2 NA 131.863 NA 0.289 NA 1.969
deg1.group.2 NA 81.971 NA 0.240 NA 1.537
deg2.group.2 NA 29.112 NA 0.139 NA 0.988
deg3.group.2 NA 6.002 NA 0.067 NA 0.361
SD(Statistic)
edges 11.113
concurrent.group1 3.581
concurrent.group2 5.092
deg0.group.1 8.031
deg1.group.1 7.987
deg2.group.1 2.838
deg3.group.1 1.835
deg0.group.2 7.122
deg1.group.2 7.233
deg2.group.2 4.785
deg3.group.2 2.349
Duration Diagnostics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 20 20.005 0.027 0.078 0.068 0.369 1.528
Dissolution Diagnostics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 0.05 0.05 0.066 0 0.219 0.001 0.017The edges seem to have a small bias, due to the relatively short duration. This is confirmed when we plot the diagnostics:
One might want to do a full tergm estimation instead of the approximation. But it is important to consider your goals, and both the relative and absolute magnitude of the bias here. For our purposes, this fit is good enough to move forward.
24.3 Epidemic Model
For the disease simulation, we will simulate an SIR epidemic in a closed population.
24.3.1 Parameterization
The epidemic model parameters are below. In this tutorial, we will use biological parameters for the infection probability per act and recovery rates as equal to demonstrate the impact of group-specific degree distributions on outcomes. Note that the group-specific parameters for the infection probability govern the risk of infection to persons in that group given contact with persons in the other group.
At the outset, 10 people in each group will be infected. For an SIR epidemic simulation, it is necessary to specify the number recovered in each group, here starting at 0.
For control settings, we will simulate 5 epidemics over 500 time steps each. Here we use the multi-core functionality in our controls.
24.3.2 Simulation
The model is simulated by inputting the fitted network model, the epidemic parameters, the initial conditions, and control settings.
Printing the model object shows the inputs and outputs from the simulation object. In particular, take note of the Variables listing; the epidemic outputs for Group 1 are listed without a suffix whereas the outputs for Group 2 are listed with a .g2 suffix.
EpiModel Simulation
=======================
Model class: netsim
Simulation Summary
-----------------------
Model type: SIR
No. simulations: 5
No. time steps: 500
No. NW groups: 2
Fixed Parameters
---------------------------
inf.prob = 0.3
rec.rate = 0.02
inf.prob.g2 = 0.3
rec.rate.g2 = 0.02
act.rate = 1
groups = 2
Model Output
-----------------------
Variables: s.num i.num r.num num s.num.g2 i.num.g2
r.num.g2 num.g2 si.flow si.flow.g2 ir.flow ir.flow.g2
Networks: sim1 ... sim5
Transmissions: sim1 ... sim5
Formation Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means)
edges 165.0 163.797 -0.729 1.024 -1.175 2.059
nodematch.group 0.0 0.000 NaN NaN NaN 0.000
concurrent.group1 12.5 13.086 4.691 0.402 1.460 0.820
concurrent.group2 37.5 36.725 -2.067 0.364 -2.129 0.494
SD(Statistic)
edges 10.829
nodematch.group 0.000
concurrent.group1 3.593
concurrent.group2 4.726
Duration Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 20 20.136 0.678 0.156 0.868 0.368 1.458
Dissolution Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 0.05 0.05 -0.716 0 -1.094 0.001 0.01724.3.3 Analysis
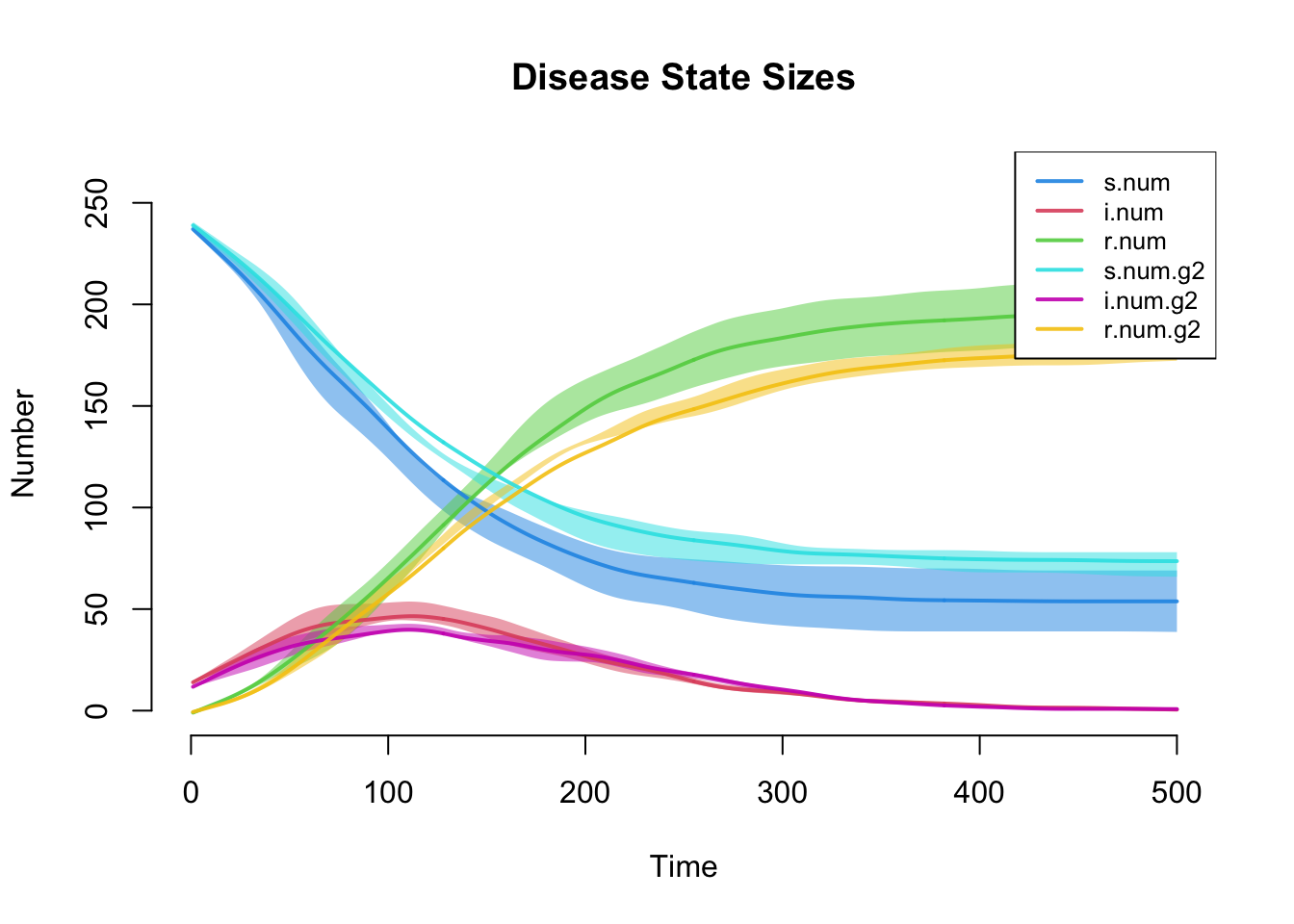
Similar to the first tutorial, plotting the netsim object shows the prevalence for each disease state in the model over time.
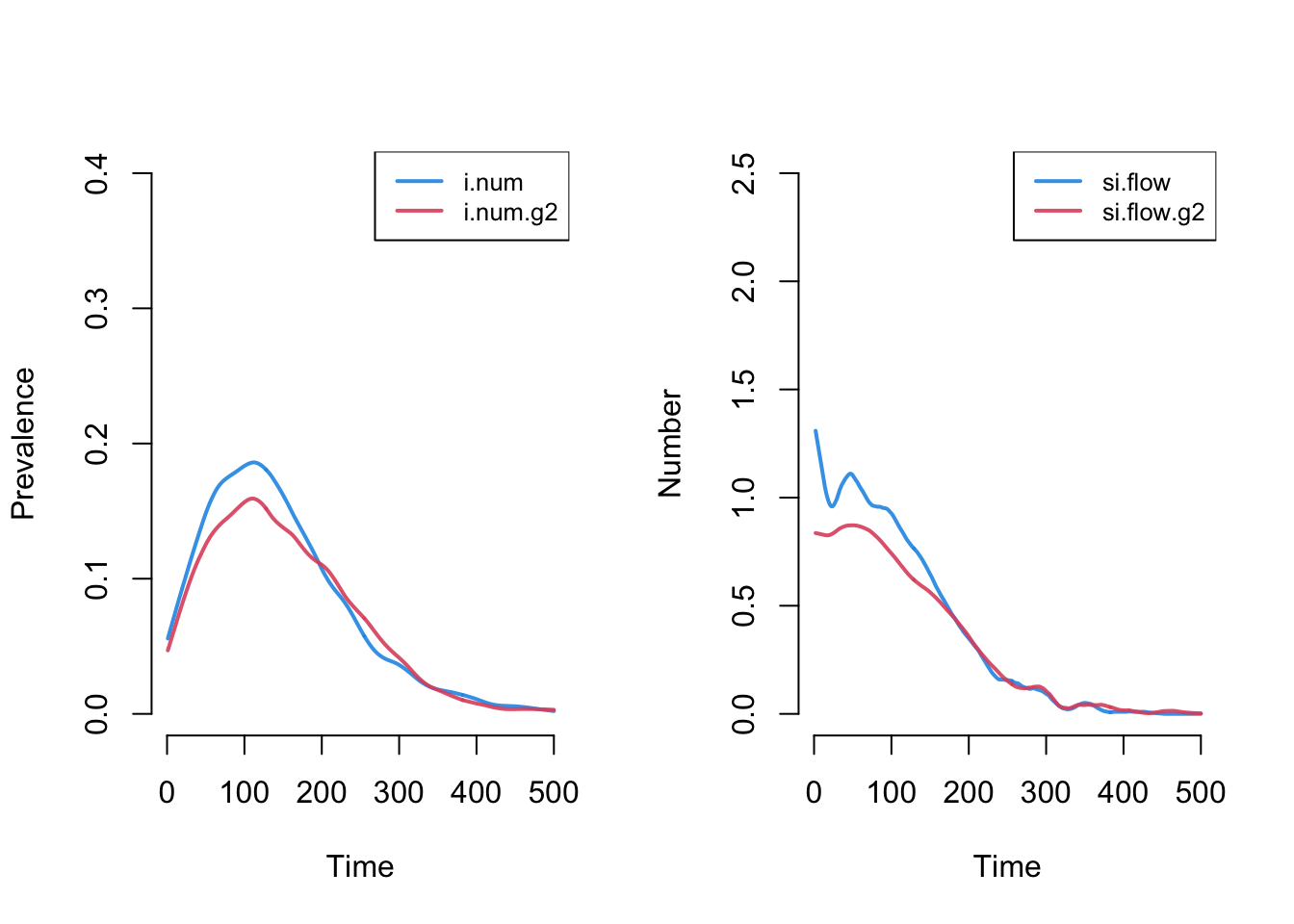
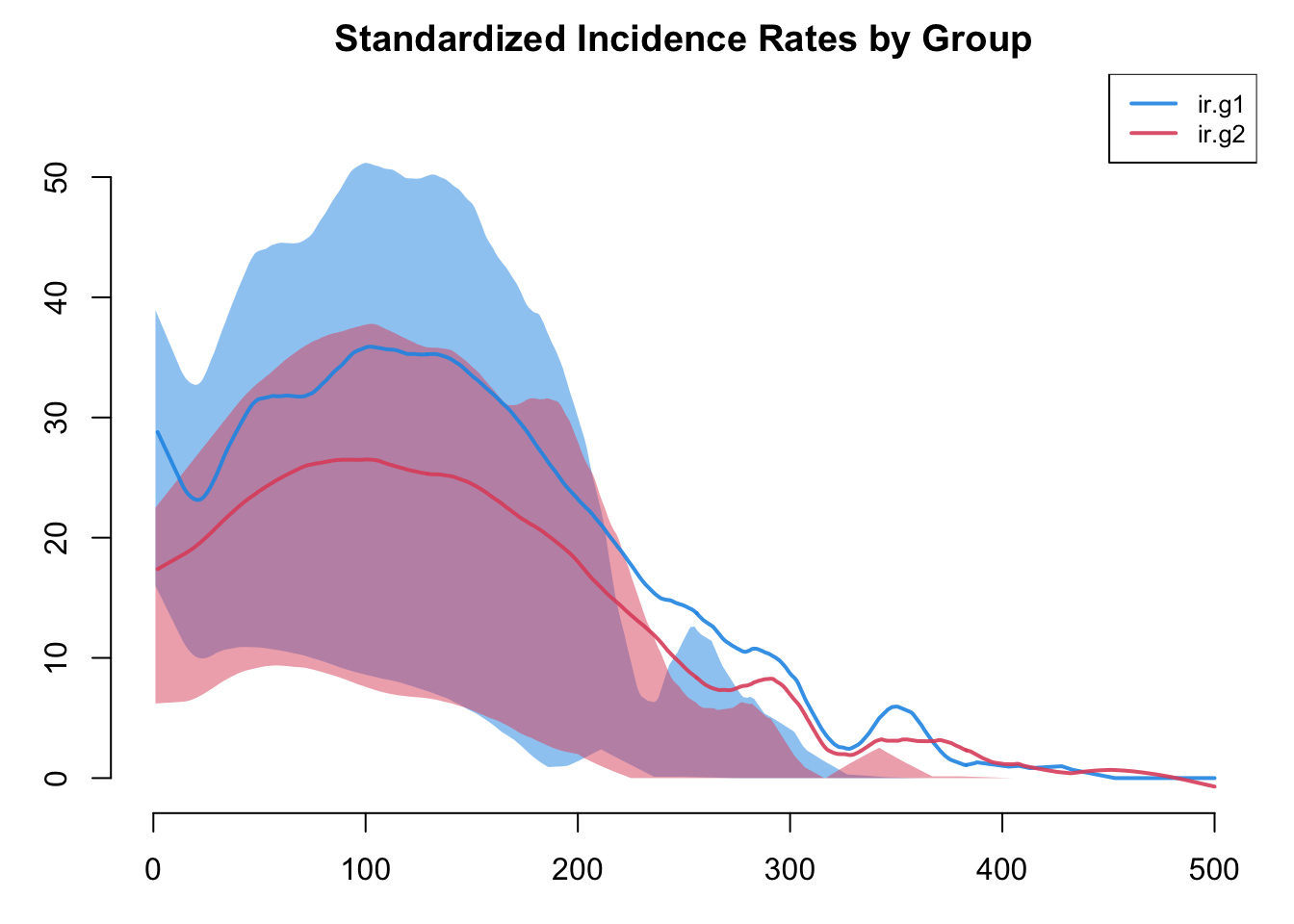
The plot below more clearly shows prevalence and incidence by group Interestingly, women (Group 1) have a slightly higher prevalence of disease during the main epidemic period, and more women than men end up in the recovered state. Men and women have the same mean degree but men have a higher prevalence of concurrency (degree 2+) than women. Following epidemiological and network theory, conditional on mean degree (which is the same for both groups in this model), concurrency increases the risk of transmission but not acquisition. So therefore, women bear the higher burden of male concurrency.
Code
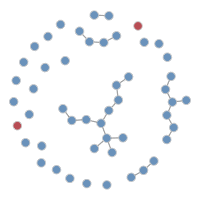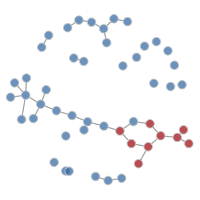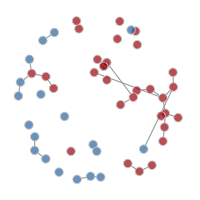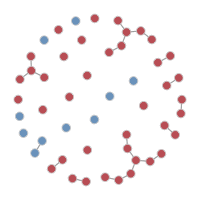
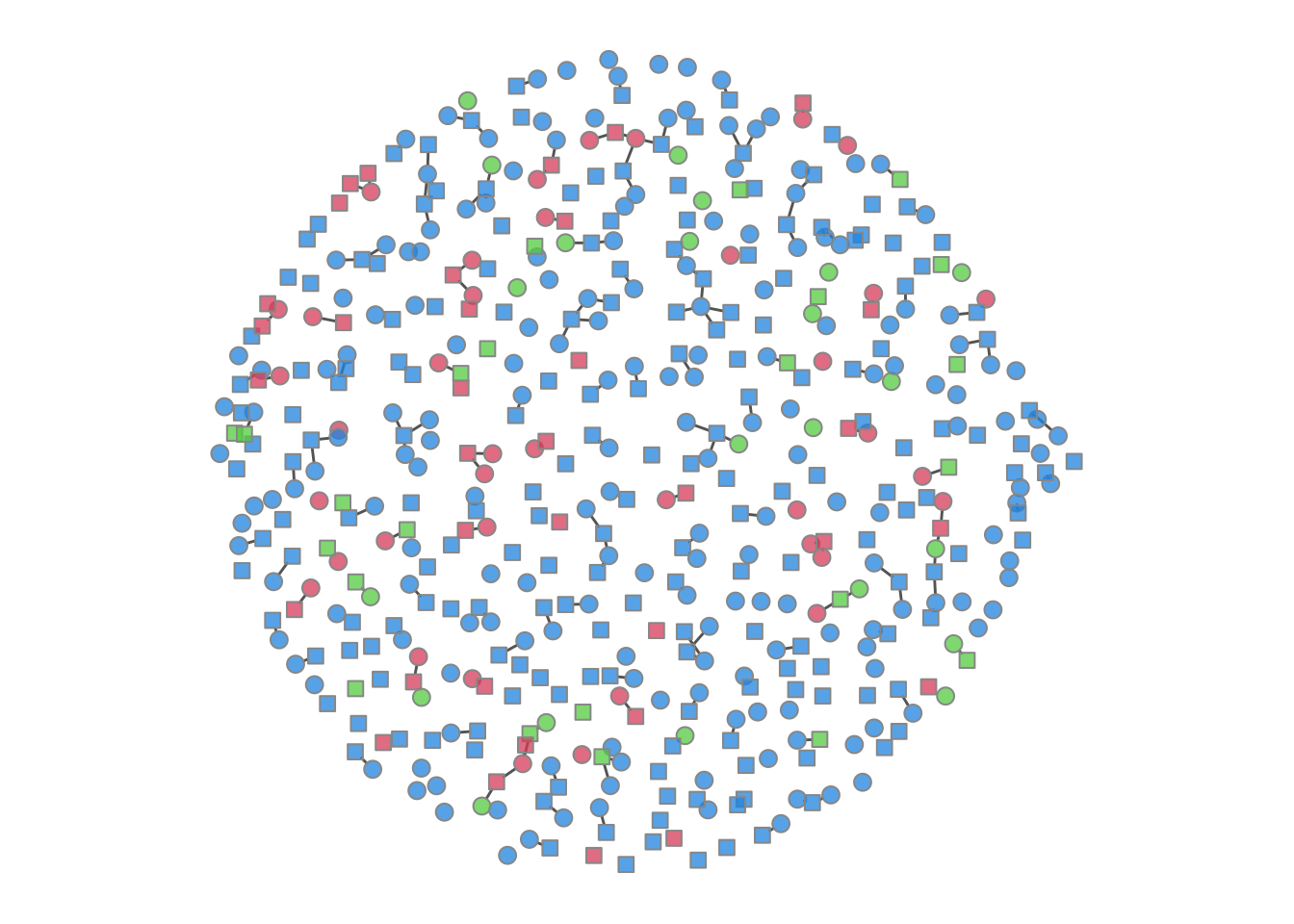
It is possible to plot the color-coded static network at various time points during the simulation, as in the last tutorial. This helps to visualize how womens’ (circle) risk is dependent on their male (square) partners’ network connections.
Code
It is possible to add new variables to an existing model object with mutate_epi, which takes inspiration from the mutate functions in the tidyverse. With a time step unit of a week, we can calculate standardized incidence rates per 100 person-years at risk using the following approach. The mutate_epi function takes a netsim object and adds a new variable.
Printing out the object, you can see that these two new variables are in the variable list.
EpiModel Simulation
=======================
Model class: netsim
Simulation Summary
-----------------------
Model type: SIR
No. simulations: 5
No. time steps: 500
No. NW groups: 2
Fixed Parameters
---------------------------
inf.prob = 0.3
rec.rate = 0.02
inf.prob.g2 = 0.3
rec.rate.g2 = 0.02
act.rate = 1
groups = 2
Model Output
-----------------------
Variables: s.num i.num r.num num s.num.g2 i.num.g2
r.num.g2 num.g2 si.flow si.flow.g2 ir.flow ir.flow.g2 ir.g1
ir.g2
Networks: sim1 ... sim5
Transmissions: sim1 ... sim5
Formation Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means)
edges 165.0 163.797 -0.729 1.024 -1.175 2.059
nodematch.group 0.0 0.000 NaN NaN NaN 0.000
concurrent.group1 12.5 13.086 4.691 0.402 1.460 0.820
concurrent.group2 37.5 36.725 -2.067 0.364 -2.129 0.494
SD(Statistic)
edges 10.829
nodematch.group 0.000
concurrent.group1 3.593
concurrent.group2 4.726
Duration Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 20 20.136 0.678 0.156 0.868 0.368 1.458
Dissolution Statistics
-----------------------
Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
edges 0.05 0.05 -0.716 0 -1.094 0.001 0.017After we add the new variable, we can plot and analyze it like any other built-in variable.
Code
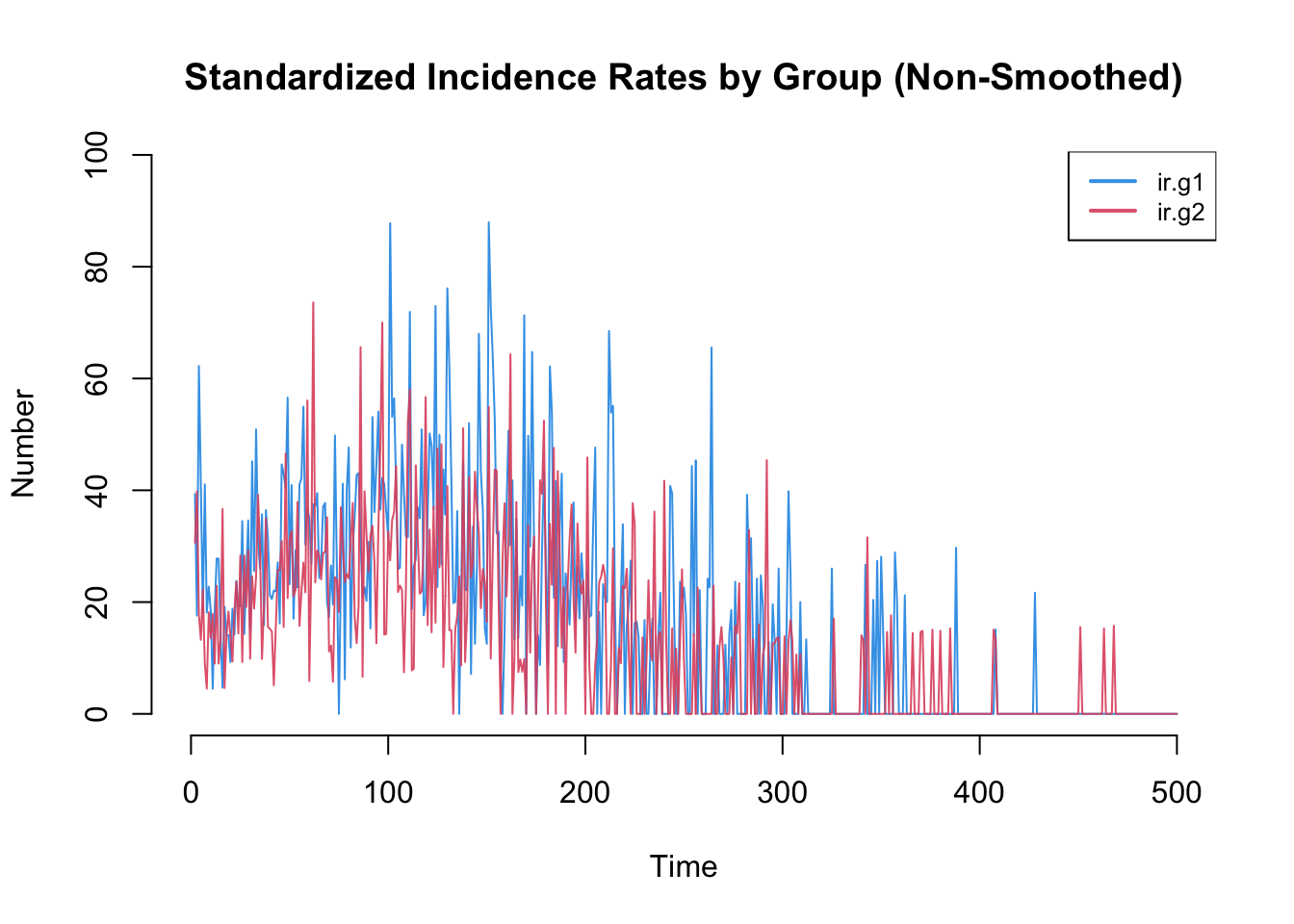
Incidence rates in particular are often highly variable. The plot by default will use a loess smoother so some of this variability may be lost. Therefore, it is worth inspecting the non-smoothed plots too.
Code
As another exercise, let’s extract the data into a data frame and then calculate some other relevant summary statistics. By default as.data.frame will extract the individual simulation data in each row, but by specifying out = "mean" we can see the time specific averages for all the variables across simulations. The variables in this data frame include both the original variables from netsim plus the two new ones we added with mutate_epi.
time s.num i.num r.num num s.num.g2 i.num.g2 r.num.g2 num.g2 si.flow
1 1 240.0 10.0 0.0 250 240.0 10.0 0.0 250 NaN
2 2 238.2 11.6 0.2 250 238.6 11.4 0.0 250 1.8
3 3 237.4 12.2 0.4 250 236.8 13.2 0.0 250 0.8
4 4 234.6 14.8 0.6 250 236.0 14.0 0.0 250 2.8
5 5 232.8 16.2 1.0 250 235.4 14.6 0.0 250 1.8
6 6 232.0 17.0 1.0 250 234.4 15.0 0.6 250 0.8
7 7 230.2 18.8 1.0 250 234.0 15.2 0.8 250 1.8
8 8 229.4 19.6 1.0 250 233.8 15.2 1.0 250 0.8
9 9 228.4 20.0 1.6 250 233.0 15.6 1.4 250 1.0
10 10 227.6 20.2 2.2 250 232.4 16.0 1.6 250 0.8
si.flow.g2 ir.flow ir.flow.g2 ir.g1 ir.g2
1 NaN NaN NaN NaN NaN
2 1.4 0.2 0.0 39.34648 30.606982
3 1.8 0.2 0.0 17.59009 39.812782
4 0.8 0.2 0.0 62.24358 17.684335
5 0.6 0.4 0.0 40.69872 13.240047
6 1.0 0.0 0.6 17.93170 22.224826
7 0.4 0.0 0.2 41.04502 8.927203
8 0.2 0.0 0.2 18.10826 4.521739
9 0.8 0.6 0.4 22.75234 18.166285
10 0.6 0.6 0.2 18.45234 13.586502One interesting summary statistic for an SIR epidemic is the cumulative incidence. Here are the mean cumulative incidences in the first and second groups across simulations. It is necessary here to use na.rm = TRUE to calculate these statistics by removing the first row where the value is undefined.
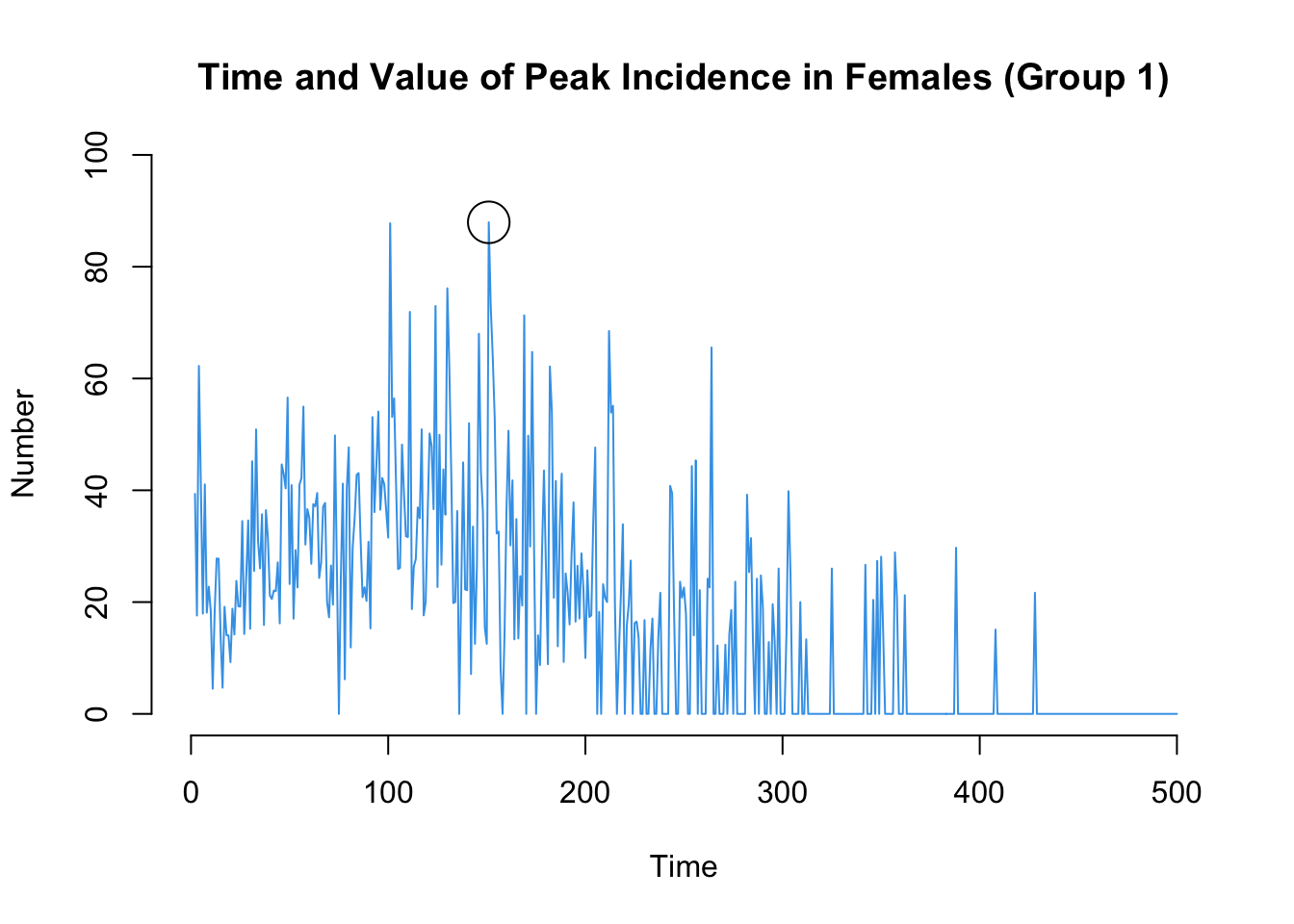
Here are two more that may be of interest: the time point of the peak standardized incidence rates, and also the value of the incidence rate at that time point.
Code
The transmission matrix again shows the individual transmission events that occurred at each time step, along with information about the infecting partner that may be relevant.
# A tibble: 10 × 8
# Groups: at, sus [10]
at sus inf network infDur transProb actRate finalProb
<dbl> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 2 98 418 1 50 0.3 1 0.3
2 2 137 482 1 132 0.3 1 0.3
3 2 243 332 1 79 0.3 1 0.3
4 2 317 238 1 69 0.3 1 0.3
5 2 402 234 1 53 0.3 1 0.3
6 4 41 317 1 2 0.3 1 0.3
7 4 52 459 1 12 0.3 1 0.3
8 4 79 482 1 134 0.3 1 0.3
9 4 161 418 1 52 0.3 1 0.3
10 4 215 263 1 12 0.3 1 0.3The infDur column shows the duration of infection (in time steps) of the infecting partner at the point of transmission. For example, an event with an infDur of 1 means that the infecting partner had just become infected themselves in the prior time step. This is the proportion of transmission that occurred in that state.
One can also convert this into a phylogeny for easier visualization of these transmission chains. First, we convert the transmission matrix into a phylo class object that can then be further analyzed and visualized with the ape package. In this transmission matrix, there are multiple trees found because we seeded the infection with 20 people as initial conditions.
Here is a standard phylogram of the first four trees. Some initial seeds lead to many downstream infections whereas others do not. Is this a function of underlying contact networks of the seeded nodes, the biology or behavior of those nodes, or something else? This could all be tested!
Code
par(mfrow = c(2, 2), mar = c(0, 0, 0, 0))
plot(tmPhylo[[1]], show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo[[2]], show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo[[3]], show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo[[4]], show.node.label = TRUE, root.edge = TRUE, cex = 0.5)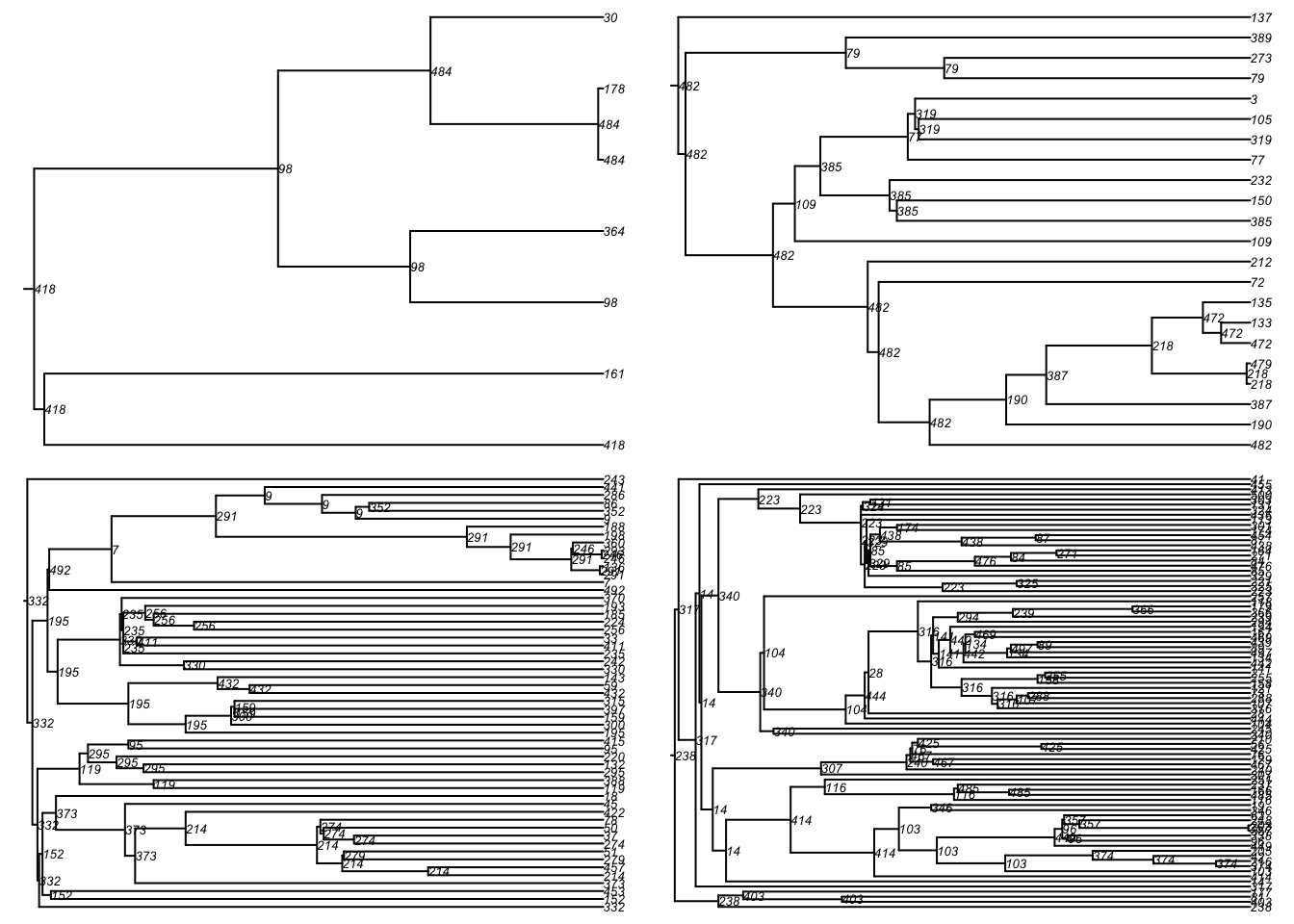
In phylo objects with multiple trees, each tree is stored with a named object. Note that these names correspond to the ID numbers in the transmission matrix itself. We can extract one tree like so:
[1] "seed_418" "seed_482" "seed_332" "seed_238" "seed_234" "seed_459"
[7] "seed_263" "seed_5" "seed_331" "seed_445" "seed_197" "seed_252"
[13] "seed_168" "seed_211" "seed_471"Then we can plot that particular tree as a phylogram and other visual types included in the ape package.
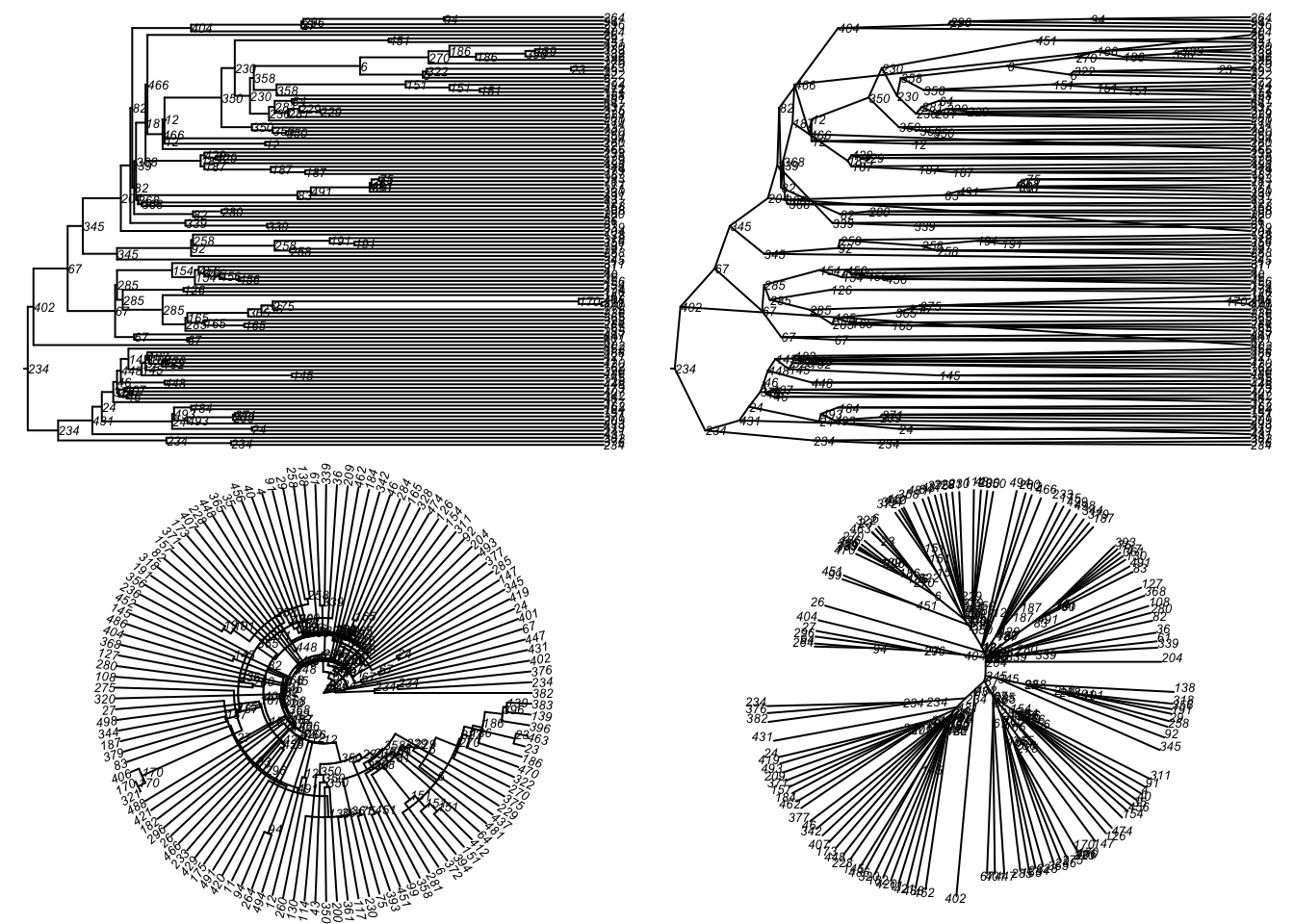
Code
par(mfrow = c(2, 2), mar = c(0, 0, 0, 0))
plot(tmPhylo5, type = "phylogram",
show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo5, type = "cladogram",
show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo5, type = "fan",
show.node.label = TRUE, root.edge = TRUE, cex = 0.5)
plot(tmPhylo5, type = "unrooted",
show.node.label = TRUE, root.edge = TRUE, cex = 0.5)