swfcalib
swfcalib.Rmdswfcalib gives a scaffold to make an automated
calibration system on a Slurm
equipped HPC. swfcalib relies on slurmworkflow
and a stored calibration object to chose which step of the calibration
to run. This allows very long calibrations to take place without
flooding the slurm queue.
For this vignette we will take as an example a simplified version of
one of our epidemic models to showcase swfcalib usage.
This model is a network model simulating HIV circulation in a population of 100k Men who have Sex with Men (MSM).
The model
A single run of our model locally looks like this:
library(EpiModelHIV)
epistats <- readRDS("data/input/epistats.rds")
netstats <- readRDS("data/input/netstats.rds")
est <- readRDS("data/input/netest.rds")
param <- param.net(
data.frame.params = read.csv("data/input/params.csv"),
netstats = netstats,
epistats = epistats
)
init <- init_msm()
control <- control_msm(
nsteps = 52 * 60,
nsims = 1,
ncores = 1
)
# The actual simulation happens here
sim <- netsim(est, param, init, control)In this example, we need to load the EpiModelHIV
package, to read 4 files located in a “data/input” folder and to pass 4
parameters to the netsim function.
The result, stored in the sim variable is not directly
usable by swfcalib.
Calibration parameters and outputs
For this example, we will focus on 9 parameters to calibrate and 9 outputs to fit to there targets.
There are many more parameters in the model. Some get their values from the literature, other free parameters to be calibrated are ignored for sake of simplicity in this example.
Outputs
The first outputs are the proportion of HIV individuals who are diagnosed (i.e. aware of their status).
Then is the proportion of HIV diagnosed who started treatment in less than a month after diagnosis.
Finally, the prevalence of diagnosed HIV in the population is our final output of interest.
As our population is race stratified between, black, hispanic and white, we get three time the number of outputs.
| Output Name | Description | Target Value |
|---|---|---|
| cc.dx.B | Portion of HIV infected who are diagnosed (black) | 0.847 |
| cc.dx.H | Portion of HIV infected who are diagnosed (hispanic) | 0.818 |
| cc.dx.W | Portion of HIV infected who are diagnosed (white) | 0.862 |
| cc.linked1m.B | Linkage to care within 1 month (black) | 0.829 |
| cc.linked1m.H | Linkage to care within 1 month (hispanic) | 0.898 |
| cc.linked1m.W | Linkage to care within 1 month (white) | 0.881 |
| i.prev.dx.B | HIV diagnosed prevalence (black) | 0.33 |
| i.prev.dx.H | HIV diagnosed prevalence (hispanic) | 0.127 |
| i.prev.dx.W | HIV diagnosed prevalence (W) | 0.084 |
Parameters
9 parameters will be calibrated to fit the model to these target values:
- The weekly probability of being tested for HIV.
- The weekly probability of starting treatment if diagnosed for HIV.
- A transmission scaler which is a parameter that encompass all the mechanism affecting transmission that are not explicitly defined in the model.
| Parameter Name | Description |
|---|---|
| hiv.test.rate_1 | Weekly test rate (black) |
| hiv.test.rate_2 | Weekly test rate (hispanic) |
| hiv.test.rate_3 | Weekly test rate (white) |
| tx.init.rate_1 | Weekly treatment start rate (black) |
| tx.init.rate_2 | Weekly treatment start rate (hispanic) |
| tx.init.rate_3 | Weekly treatment start rate (white) |
| hiv.trans.scale_1 | Transmission scaler (black) |
| hiv.trans.scale_2 | Transmission scaler (hispanic) |
| hiv.trans.scale_3 | Transmission scaler (white) |
Relationship between parameters and outputs
Simply by looking at them, we can see that all the parameters do not relate to all the outputs in the same way.
The hiv.test.rate parameters directly relate to the
cc.dx outputs (proportion) of diagnosed. Also, no other
parameter in our list would affect these outcomes. Therefore, we have 3
one to one relationship. For each race, the test rate will govern the
diagnosed proportion.
A similar situation can be described for linkage to care
(cc.linked1m) and treatment start rate
tx.init.rate. (the denominator is the number of diagnosed
individuals, making it independent of cc.dx).
The prevalence (i.prev.dx), on the other hand, depends
on all 9 parameters. The proportion of diagnosed influences the number
of treated which then influences the number of individuals able to
transmit. The scalers then affect directly the transmission for one
population. But as they modify the prevalence, they also indirectly
influence the transmission in the other sub populations.
However, once the hiv.test.rate and
tx.init.rate are calibrated and their values fixed,
i.prev.dx only depends on the hiv.trans.scale
parameters. Reducing a 9 parameters 3 outputs problem into a simpler 3
parameters, 3 outputs one.
The following graph illustrate these relationships.
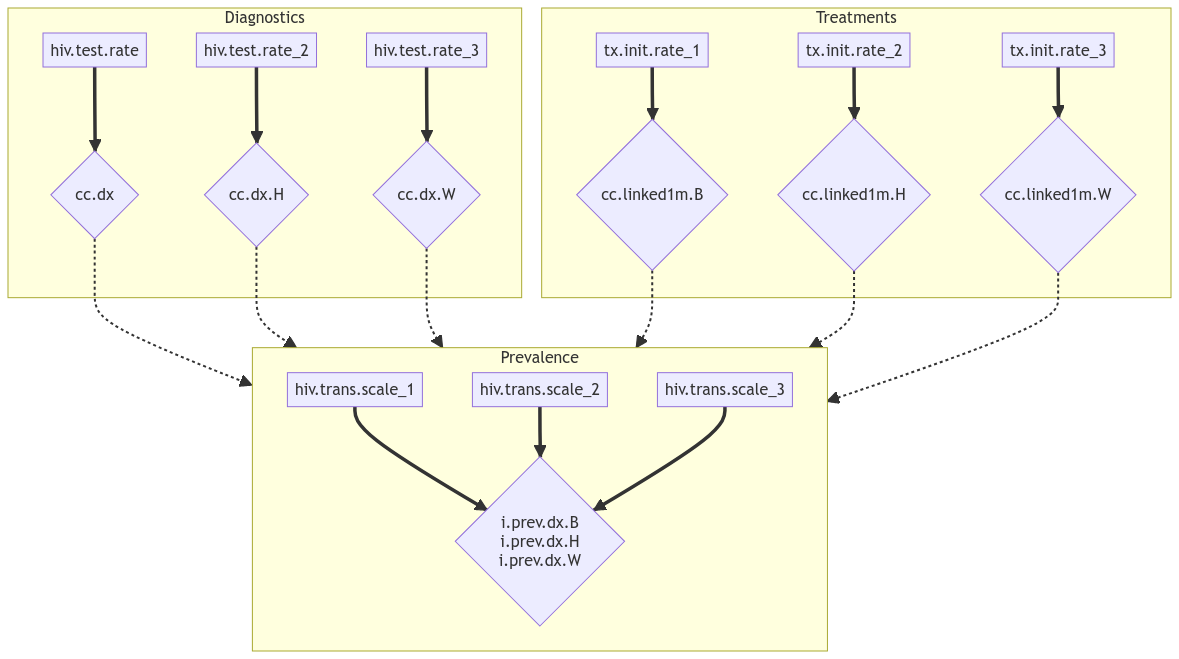
Calibration structure
Knowing these parameters - outputs relationship, we can define a calibration structure to minimize the number of simulations required.
A first wave will calibrate simultaneously the 3
hiv.test.rate and the 3 tx.init.rate
parameters. This means that each run of the simulation will test a value
for each of these 6 parameters. This idea is similar factorial
experiment design and is only possible because of the independence
between the parameters and their respective outcomes.
After these 6 parameters are calibrated (i.e. given fixed and final
values), the hiv.trans.scale parameters will be calibrated
to fit the i.prev.dx outcomes to their target values.
Waves and jobs
To express this structure, we need to define calibration jobs that are going to be run in parallel within several waves.
In our example we will have two waves, the first one for the
hiv.test.rate and tx.init.rate parameters and
a second for the hiv.trans.scale parameters.
Jobs
Formally, swfcalib defines a job as a set of
parameters to be calibrated by trying to make a set of
outcomes reach a set of targets.
Each job needs a function to make the next set of parameter proposals to test as well as a function for checking if the proposals gave sufficiently good results. This latter function is in charge of stopping the calibration process for the current job.
Waves
A wave is a set of multiple jobs that can be run in parallel (i.e. independent from one another).
In practice, swfcalib takes the proposals from all the
jobs in a wave, combine them and run one simulation per proposal. If you
have a 3 job wave, each making 10 proposal, only 10 simulations
will be run. On the evaluation step each job will only assess the
quality of it’s own outcomes.
Once all the jobs in a wave are finished, the system moves to the next one if any, using the parameters calibrated on the previous ones.
Implementing the model function
Now that we have a good conceptual idea how the calibration will go,
we need to make our code compatible with swfcalib.
For the model function it is pretty straightforward. We need a function with this signature:
model <- function(proposal) {
# simulation code
return(results)
}Where proposal is an one row tibble with
each column being a parameter to calibrate. In our case,
proposal is a 1 row 9 columns tibble.
And results is a one row tibble where each
column is an output for the calibration process. In this case,
results must also be a 1 row 9 columns
tibble.
Below is the code for our example.
model <- function(proposal) {
# Load all required elements
library(EpiModelHIV)
library(dplyr)
epistats <- readRDS("data/input/epistats.rds")
netstats <- readRDS("data/input/netstats.rds")
est <- readRDS("data/input/netest.rds")
param <- param.net(
data.frame.params = read.csv("data/input/params.csv"),
netstats = netstats,
epistats = epistats
)
init <- init_msm()
control <- control_msm(
nsteps = 52 * 60,
nsims = 1,
ncores = 1
)
# Proposal to scenario -------------------------------------------------------
scenario <- EpiModelHPC::swfcalib_proposal_to_scenario(proposal)
param_sc <- EpiModel::use_scenario(param, scenario)
# Run the simulation ---------------------------------------------------------
sim <- netsim(est, param_sc, init, control)
# Process the results -------------------------------------------------------
results <- as_tibble(sim) |>
mutate_calibration_targets() |>
filter(time >= max(time) - 52) |>
select(
cc.dx.B, cc.dx.H, cc.dx.W,
cc.linked1m.B, cc.linked1m.H, cc.linked1m.W,
i.prev.dx.B, i.prev.dx.H, i.prev.dx.W,
) |>
summarise(across( everything(), ~ mean(.x, na.rm = TRUE)))
# Return the one row `tibble`
return(results)
}Several things are of importance here:
- We call
librarywithin the function. It is not usual, but we must remember that this will be run on ansbatchjob from a clean environment. Therefore, the function must load all the required libraries and files. - We must adapt the
proposalto be used by the simulation. In this case, a helper functionswfcalib_proposal_to_scenariodoes it. But simply copying the values from theproposaltibbleworks as well. - After the simulation, the outputs must be processed to produce a
correctly formatted
tibble. In this case, we take the mean over the last 52 weeks (1 year) for each of the desired outputs.
As the users of swfcalib, we are responsible for the
correct output of model.
Also, model must be a function that run a single
simulation and produce a single set of results. This
function will be run in parallel to test several proposals at once.
Configuring an swfcalib system
swfcalib maintain a calib_object that store
the state of the calibration as well as all it needs to operate. This
object is first defined locally and then updated on the HPC as the
calibration progresses.
This object is an R list with 3 elements:
state, config and waves.
We won’t cover state now as it is created by
swfcalib and only edited by it.
Configuration
config is a list with the following
elements:
-
simulator: the function to calibrationmodelin our case -
root_directory: where the system will store itself on the HPC -
n_sims: the number of simulations to run in parallel at each iteration -
max_iteration: the maximum number of iteration before stopping the calibration if a satisfactory state is not found. This is a fail-safe mechanism to avoid consuming HPC resources when the calibration does not work. -
default_proposal: default values for the calibrated parameters. This fixes the values for the parameters that are to be calibrated in later waves. Once a job is finished, the calibrated value is stored here and used for the next runs
config = list(
simulator = model,
root_directory = "data/calib",
n_sims = n_sims,
max_iteration = 100,
default_proposal = dplyr::tibble(
hiv.test.rate_1 = 0.004123238,
hiv.test.rate_2 = 0.003771226,
hiv.test.rate_3 = 0.005956663,
tx.init.rate_1 = 0.2981623,
tx.init.rate_2 = 0.3680919,
tx.init.rate_3 = 0.358254,
hiv.trans.scale_1 = 2.470962,
hiv.trans.scale_2 = 0.4247816,
hiv.trans.scale_3 = 0.3342994
)
)Calibration jobs
A calibration job is a list with 6
elements:
-
targets: the name of the outputs to fit (>= 1) -
targets_val: the target values -
params: the names of the parameters to calibrate (>=1) -
initial_proposals: atibblewith the values to be tested for the first run of the simulation. -
make_next_proposals: a function that will define which proposals to make next -
get_result: a function to define is the calibration is done for this job
We will get into the details of these elements later on.
Below is an example of the job for calibrating the
hiv.trans.scale parameters using the i.prev.dx
outputs.
job1 = list(
targets = paste0("i.prev.dx.", c("B", "H", "W")),
targets_val = c(0.33, 0.127, 0.09),
params = paste0("hiv.trans.scale_", 1:3),
initial_proposals = dplyr::tibble(
hiv.trans.scale_1 = sample(seq(1, 4, length.out = n_sims)),
hiv.trans.scale_2 = sample(seq(0.2, 0.6, length.out = n_sims)),
hiv.trans.scale_3 = sample(seq(0.2, 0.6, length.out = n_sims))
),
make_next_proposals =
swfcalib::make_proposer_se_range(n_sims, retain_prop = 0.3),
get_result = swfcalib::determ_end_thresh(
thresholds = rep(0.02, 3),
n_enough = 100
)
)Complete configuration
Below is the complete calib_object defined locally. Note
that we define an n_sims variable at the beginning and
reuse it all over the configuration to ensure that we have the correct
number of proposals at each step.
n_sims <- 400
calib_object <- list(
config = list(
simulator = model,
default_proposal = dplyr::tibble(
hiv.test.rate_1 = 0.004123238,
hiv.test.rate_2 = 0.003771226,
hiv.test.rate_3 = 0.005956663,
tx.init.rate_1 = 0.2981623,
tx.init.rate_2 = 0.3680919,
tx.init.rate_3 = 0.358254,
hiv.trans.scale_1 = 2.470962,
hiv.trans.scale_2 = 0.4247816,
hiv.trans.scale_3 = 0.3342994
),
root_directory = "data/calib",
max_iteration = 100,
n_sims = n_sims
),
waves = list(
wave1 = list(
job1 = list(
targets = "cc.dx.B",
targets_val = 0.847,
params = c("hiv.test.rate_1"), # target: 0.00385
initial_proposals = dplyr::tibble(
hiv.test.rate_1 = seq(0.002, 0.006, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job2 = list(
targets = "cc.dx.H",
targets_val = 0.818,
params = c("hiv.test.rate_2"), # target: 0.0038
initial_proposals = dplyr::tibble(
hiv.test.rate_2 = seq(0.002, 0.006, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job3 = list(
targets = "cc.dx.W",
targets_val = 0.862,
params = c("hiv.test.rate_3"), # target: 0.0069
initial_proposals = dplyr::tibble(
hiv.test.rate_3 = seq(0.004, 0.008, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job4 = list(
targets = paste0("cc.linked1m.", c("B", "H", "W")),
targets_val = c(0.829, 0.898, 0.881),
params = paste0("tx.init.rate_", 1:3),
initial_proposals = dplyr::tibble(
tx.init.rate_1 = sample(seq(0.1, 0.5, length.out = n_sims)),
tx.init.rate_2 = sample(tx.init.rate_1),
tx.init.rate_3 = sample(tx.init.rate_1),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 3)
)
),
wave2 = list(
job1 = list(
targets = paste0("i.prev.dx.", c("B", "H", "W")),
targets_val = c(0.33, 0.127, 0.09),
params = paste0("hiv.trans.scale_", 1:3),
initial_proposals = dplyr::tibble(
hiv.trans.scale_1 = sample(seq(1, 4, length.out = n_sims)),
hiv.trans.scale_2 = sample(seq(0.2, 0.6, length.out = n_sims)),
hiv.trans.scale_3 = sample(seq(0.2, 0.6, length.out = n_sims))
),
make_next_proposals =
swfcalib::make_proposer_se_range(n_sims, retain_prop = 0.3),
get_result = swfcalib::determ_end_thresh(
thresholds = rep(0.02, 3),
n_enough = 100
)
)
)
)
)Proposers and calibration check
As mentioned earlier, each calibration job needs a function
to define which proposal to make at the next iteration
(make_next_proposals) and a function to assess if the
calibration is finished (get_result).
We won’t get into the details of how these functions work but focus instead on their practical use. You can have a look at their respective documentation for more details.
HIV test rate
The 3 jobs related to hiv.test.rate and
proportion of diagnosed among infected (cc.dx) use the same
approach:
The proposer function is generated by a function
factory: make_shrink_proposer(n_sims, shrink = 2). This
proposer will shrink the range of proposals by a factor 2 around the
best guess so far.
The calibration assessor is made by
determ_poly_end(0.001, poly_n = 5). This function uses
linear model with a degree 5 polynomial to predict the best value for
the parameter. The calibration is considered finished when the predicted
value is less than threshold away from the target (here
0.001) AND when the prediction is not improving after the
last iteration.
Linkage to care
Linkage to care uses the same functions but all 3 parameters and targets are fitted at once. This here implies that a single model is fitted for all the data. It works very well in this specific case as the relationship between linkage to care and treatment uptake rate is consistent over the three groups.
HIV prevalence
HIV prevalence and transmission scale is harder to calibrate as the 3 parameters and outputs are linked together.
At each iteration the squared error over all 3 targets is calculated
for each proposal. The ranges for the next round of proposals are the
ranges of these best 30% proposals of the previous rounds. The function
factory here is make_proposer_se_range. It takes a
retain_prop argument that governs which proportion of the
simulations are used to define the new ranges.
The calibration is considered done when 100 simulations where the 3
outputs are less than thresholds away from there respective
targets. The determ_end_thresh function factory allow us to
define the number of good simulations required and the
thresholds for each output.
Running a calibration system
Now that all the pieces are ready, we can setup the workflow using
the slurmworkflow
package.
This will allow the HPC to run the calibration steps in a loop until the calibration is finished.
The following script setup a complete calibration workflow with what was defined so far.
The main thing you may want to change in this script is the
batch_size variable that governs how many sims are run in
parallel on a single cluster. On the RSPH HPC we set it to 8. Therefore,
50 batches of 8 are submitted to slurm at once to make the
400 simulations requested per iteration.
A mail is sent if a job fails or at the end of the 3rd calibration step. This latter indicate that the calibration process is done.
library("slurmworkflow")
# Define the `model` function
model <- function(proposal) {
# Load all required elements
library(EpiModelHIV)
library(dplyr)
epistats <- readRDS("data/input/epistats.rds")
netstats <- readRDS("data/input/netstats.rds")
est <- readRDS("data/input/netest.rds")
param <- param.net(
data.frame.params = read.csv("data/input/params.csv"),
netstats = netstats,
epistats = epistats
)
init <- init_msm()
control <- control_msm(
nsteps = 52 * 60,
nsims = 1,
ncores = 1
)
# Proposal to scenario -------------------------------------------------------
scenario <- EpiModelHPC::swfcalib_proposal_to_scenario(proposal)
param_sc <- EpiModel::use_scenario(param, scenario)
# Run the simulation ---------------------------------------------------------
sim <- netsim(est, param_sc, init, control)
# Process the results -------------------------------------------------------
results <- as_tibble(sim) |>
mutate_calibration_targets() |>
filter(time >= max(time) - 52) |>
select(
cc.dx.B, cc.dx.H, cc.dx.W,
cc.linked1m.B, cc.linked1m.H, cc.linked1m.W,
i.prev.dx.B, i.prev.dx.H, i.prev.dx.W,
) |>
summarise(across( everything(), ~ mean(.x, na.rm = TRUE)))
# Return the one row `tibble`
return(results)
}
# Create the `calib_object`
n_sims <- 400
calib_object <- list(
config = list(
simulator = model,
default_proposal = dplyr::tibble(
hiv.test.rate_1 = 0.004123238,
hiv.test.rate_2 = 0.003771226,
hiv.test.rate_3 = 0.005956663,
tx.init.rate_1 = 0.2981623,
tx.init.rate_2 = 0.3680919,
tx.init.rate_3 = 0.358254,
hiv.trans.scale_1 = 2.470962,
hiv.trans.scale_2 = 0.4247816,
hiv.trans.scale_3 = 0.3342994
),
root_directory = "data/calib",
max_iteration = 100,
n_sims = n_sims
),
waves = list(
wave1 = list(
job1 = list(
targets = "cc.dx.B",
targets_val = 0.847,
params = c("hiv.test.rate_1"), # target: 0.00385
initial_proposals = dplyr::tibble(
hiv.test.rate_1 = seq(0.002, 0.006, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job2 = list(
targets = "cc.dx.H",
targets_val = 0.818,
params = c("hiv.test.rate_2"), # target: 0.0038
initial_proposals = dplyr::tibble(
hiv.test.rate_2 = seq(0.002, 0.006, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job3 = list(
targets = "cc.dx.W",
targets_val = 0.862,
params = c("hiv.test.rate_3"), # target: 0.0069
initial_proposals = dplyr::tibble(
hiv.test.rate_3 = seq(0.004, 0.008, length.out = n_sims),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 5)
),
job4 = list(
targets = paste0("cc.linked1m.", c("B", "H", "W")),
targets_val = c(0.829, 0.898, 0.881),
params = paste0("tx.init.rate_", 1:3),
initial_proposals = dplyr::tibble(
tx.init.rate_1 = sample(seq(0.1, 0.5, length.out = n_sims)),
tx.init.rate_2 = sample(tx.init.rate_1),
tx.init.rate_3 = sample(tx.init.rate_1),
),
make_next_proposals = swfcalib::make_shrink_proposer(n_sims, shrink = 2),
get_result = swfcalib::determ_poly_end(0.001, poly_n = 3)
)
),
wave2 = list(
job1 = list(
targets = paste0("i.prev.dx.", c("B", "H", "W")),
targets_val = c(0.33, 0.127, 0.09),
params = paste0("hiv.trans.scale_", 1:3),
initial_proposals = dplyr::tibble(
hiv.trans.scale_1 = sample(seq(1, 4, length.out = n_sims)),
hiv.trans.scale_2 = sample(seq(0.2, 0.6, length.out = n_sims)),
hiv.trans.scale_3 = sample(seq(0.2, 0.6, length.out = n_sims))
),
make_next_proposals =
swfcalib::make_proposer_se_range(n_sims, retain_prop = 0.3),
get_result = swfcalib::determ_end_thresh(
thresholds = rep(0.02, 3),
n_enough = 100
)
)
)
)
)
# Workflow ---------------------------------------------------------------------
# Use preconfigured HPC settings
hpc_configs <- EpiModelHPC::swf_configs_rsph(
partition = "epimodel",
r_version = "4.3",
mail_user = "<your_user>@emory.edu"
)
wf <- create_workflow(
wf_name = "Vignette_auto_calib",
default_sbatch_opts = hpc_configs$default_sbatch_opts
)
# Calibration step 1
wf <- add_workflow_step(
wf_summary = wf,
step_tmpl = step_tmpl_do_call(
what = swfcalib::calibration_step1,
args = list(
n_cores = 8,
calib_object = calib_object
),
setup_lines = hpc_configs$r_loader
),
sbatch_opts = list(
"cpus-per-task" = 8,
"time" = "00:20:00",
"mem-per-cpu" = "4G",
"mail-type" = "FAIL"
)
)
# Calibration step 2
batch_size <- 8
batch_numbers <- swfcalib:::get_batch_numbers(calib_object, batch_size)
wf <- add_workflow_step(
wf_summary = wf,
step_tmpl = step_tmpl_map(
FUN = swfcalib::calibration_step2,
batch_num = batch_numbers,
setup_lines = hpc_configs$r_loader,
max_array_size = 500,
MoreArgs = list(
n_cores = batch_size,
n_batches = max(batch_numbers),
calib_object = calib_object
)
),
sbatch_opts = list(
"cpus-per-task" = batch_size,
"time" = "05:00:00",
"mem-per-cpu" = "5G",
"mail-type" = "FAIL"
)
)
# Calibration step 3
wf <- add_workflow_step(
wf_summary = wf,
step_tmpl = step_tmpl_do_call(
what = swfcalib::calibration_step3,
args = list(
calib_object = calib_object
),
setup_lines = hpc_configs$r_loader
),
sbatch_opts = list(
"cpus-per-task" = 1,
"time" = "00:20:00",
"mem-per-cpu" = "8G",
"mail-type" = "END"
)
)