Runs diagnostic simulations on an ERGM/STERGM estimated with
netest to assess whether the fitted model reproduces the
intended network features. Both static (cross-sectional) and
dynamic (temporal) diagnostics are supported. This is the
recommended second step in the network modeling pipeline, after
estimation with netest and before epidemic simulation with
netsim.
Usage
netdx(
x,
nsims = 1,
dynamic = TRUE,
nsteps = NULL,
nwstats.formula = "formation",
set.control.ergm = control.simulate.formula(),
set.control.tergm = control.simulate.formula.tergm(MCMC.maxchanges =
.Machine$integer.max),
sequential = TRUE,
keep.tedgelist = FALSE,
keep.tnetwork = FALSE,
verbose = TRUE,
ncores = 1,
skip.dissolution = FALSE,
future.use.plan = FALSE
)Arguments
- x
An
EpiModelobject of classnetest.- nsims
Number of simulations to run. For dynamic diagnostics, 5–10 simulations are usually sufficient to assess model fit. For static diagnostics, use 10,000+ draws to obtain stable estimates.
- dynamic
If
TRUE, runs dynamic diagnostics that simulate the temporal network forward in time, checking both formation targets and partnership duration/dissolution. IfFALSE, draws from the static ERGM fit to check cross-sectional network structure only (faster, but does not verify dissolution dynamics). Static diagnostics are only available when the model was fit with the edges dissolution approximation (edapprox = TRUEinnetest).- nsteps
Number of time steps per simulation (dynamic simulations only). Should be at least several multiples of the longest target partnership duration to allow the duration and dissolution statistics to stabilize. For example, if the target duration is 50, running for 500 time steps is a reasonable starting point.
- nwstats.formula
A right-hand sided ERGM formula with the network statistics of interest. The default is the formation formula of the network model contained in
x. You may track additional network statistics beyond the formation terms by specifying them here, such as~ edges + meandeg + concurrent + degree(0:4). This is useful for verifying that the model produces reasonable values for network features that were not directly targeted in the formation model.- set.control.ergm
Control arguments passed to
ergm'ssimulate_formula.network(see details).- set.control.tergm
Control arguments passed to
tergm'ssimulate_formula.network(see details).- sequential
For static diagnostics (
dynamic=FALSE): ifFALSE, each of thensimssimulated Markov chains begins at the initial network; ifTRUE, the end of one simulation is used as the start of the next.- keep.tedgelist
If
TRUE, keep the timed edgelist generated from the dynamic simulations. Returned in the form of a list of matrices, with one entry per simulation. Accessible at$edgelist.- keep.tnetwork
If
TRUE, keep the full networkDynamic objects from the dynamic simulations. Returned in the form of a list of nD objects, with one entry per simulation. Accessible at$network.- verbose
If
TRUE, print progress to the console.- ncores
Number of processor cores to run multiple simulations on, using the
futureframework.- skip.dissolution
If
TRUE, skip over the calculations of duration and dissolution stats innetdx.- future.use.plan
If
TRUE,netsimwill use the user-definedfuture::planfor its parallelization. Otherwise,multisessionis used withworkers = ncores.
Value
A list of class netdx. Use print() to view summary tables of
formation statistics, duration, and dissolution diagnostics. Use
plot.netdx to visualize these diagnostics over time. Use
as.data.frame.netdx() to extract timed edgelists (if
keep.tedgelist = TRUE).
Details
The netdx function handles dynamic network diagnostics for network
models fit with the netest function. Given the fitted model,
netdx simulates a specified number of dynamic networks for a specified
number of time steps per simulation. The network statistics in
nwstats.formula are saved for each time step. Summary statistics for
the formation model terms, as well as dissolution model and relational
duration statistics, are then calculated and can be accessed when printing or
plotting the netdx object. See print.netdx and plot.netdx
for details on printing and plotting.
Control Arguments
Models fit with the full STERGM method in netest (setting the
edapprox argument to FALSE) require only a call to
tergm's simulate_formula.network. Control parameters for those
simulations may be set using set.control.tergm in netdx.
The parameters should be input through the control.simulate.formula.tergm
function, with the available parameters listed in the
tergm::control.simulate.formula.tergm help page in the tergm
package.
Models fit with the ERGM method with the edges dissolution approximation
(setting edapprox to TRUE) require a call first to
ergm's simulate_formula.network for simulating an initial
network, and second to tergm's simulate_formula.network for
simulating that static network forward through time. Control parameters may
be set for both processes in netdx. For the first, the parameters
should be input through the control.simulate.formula() function, with
the available parameters listed in the
ergm::control.simulate.formula help
page in the ergm package. For the second, parameters should be input
through the control.simulate.formula.tergm() function, with the
available parameters listed in the tergm::control.simulate.formula.tergm
help page in the tergm package. An example is shown below.
Static vs. Dynamic Diagnostics
Static diagnostics (dynamic = FALSE) draw many independent networks from
the fitted ERGM and compare the resulting statistics to the target values.
This is fast and checks whether the cross-sectional structure is correct,
but it does not verify partnership durations or dissolution rates. Dynamic
diagnostics (dynamic = TRUE) simulate the full temporal network forward
in time, checking both formation targets and dissolution/duration dynamics.
Dynamic diagnostics are slower but more comprehensive, and are required to
verify models that will be used with vital dynamics (arrivals/departures).
Interpreting Diagnostics
After running netdx, use print() and plot.netdx to inspect the
results. Key indicators of a good model fit include:
Formation statistics: The "Sim Mean" should be close to the "Target" value. A small "Pct Diff" (< 5\ indicate good fit.
Duration statistics (dynamic only): The simulated mean edge durations should match the values passed to
dissolution_coefs.Dissolution statistics (dynamic only): The simulated dissolution rates should be approximately
1 / duration.
Common problems: If formation statistics are off, the ERGM may need
increased burn-in (via set.control.ergm), or the target statistics
may be incompatible (e.g., specifying more edges than the network can
support). If durations are off but formation is correct, verify that
d.rate was correctly specified in dissolution_coefs for models
with vital dynamics.
See also
Estimate the network model with netest before running diagnostics.
Plot diagnostics with plot.netdx and print summary tables with
print.netdx. After diagnostics confirm a good fit, simulate the
epidemic with netsim.
Examples
# Static diagnostics on a simple model
nw <- network_initialize(n = 100)
formation <- ~edges
target.stats <- 50
coef.diss <- dissolution_coefs(dissolution = ~offset(edges), duration = 25)
est <- netest(nw, formation, target.stats, coef.diss, verbose = FALSE)
#> Starting simulated annealing (SAN)
#> Iteration 1 of at most 4
#> Finished simulated annealing
#> Starting maximum pseudolikelihood estimation (MPLE):
#> Obtaining the responsible dyads.
#> Evaluating the predictor and response matrix.
#> Maximizing the pseudolikelihood.
#> Finished MPLE.
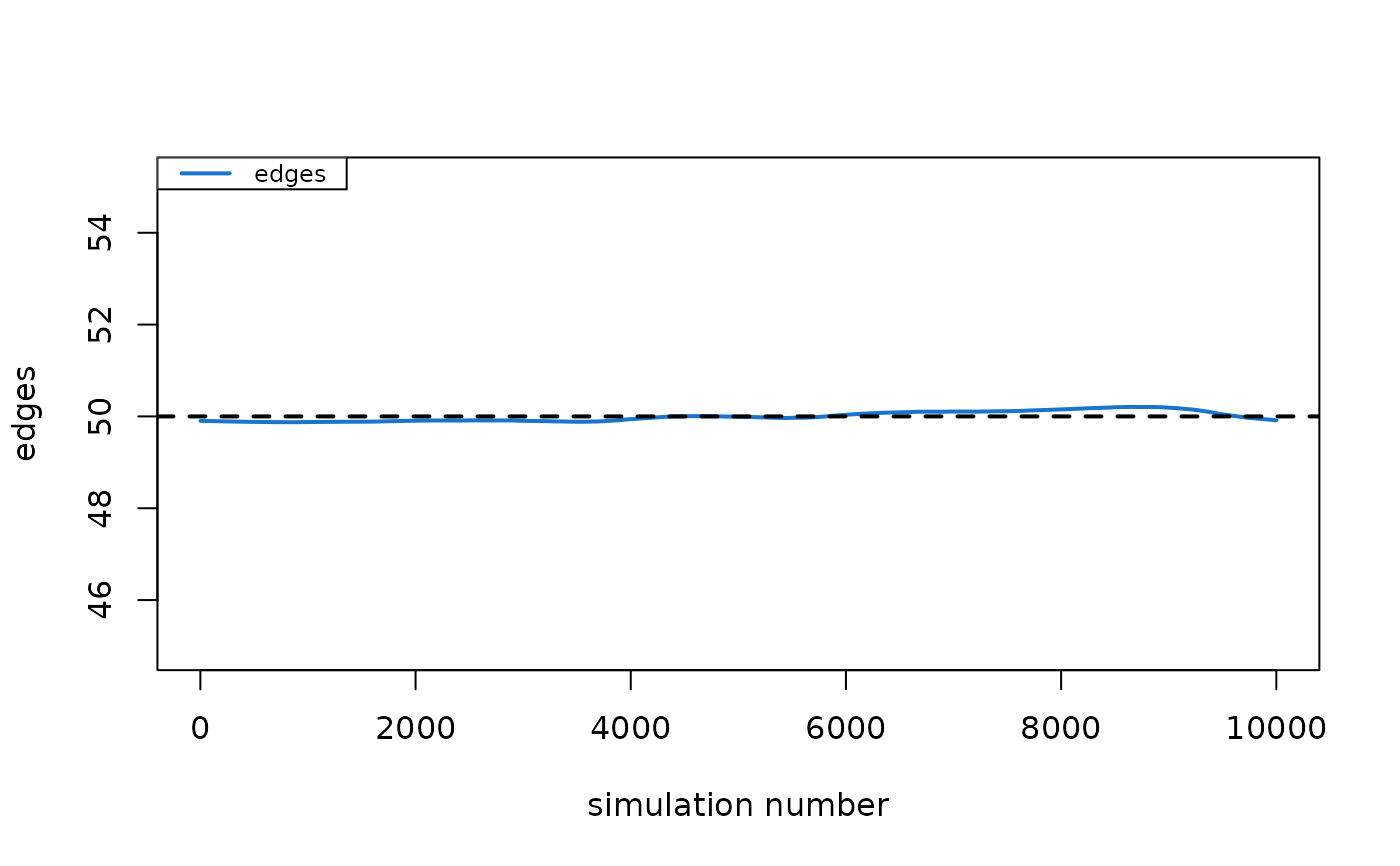
dx <- netdx(est, nsims = 1e4, dynamic = FALSE, verbose = FALSE)
#> Sampling ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ 96% | ETA: 0s
dx
#> EpiModel Network Diagnostics
#> =======================
#> Diagnostic Method: Static
#> Simulations: 10000
#>
#> Formation Diagnostics
#> -----------------------
#> Target Sim Mean Pct Diff Sim SE Z Score SD(Sim Means) SD(Statistic)
#> edges 50 50.009 0.019 0.072 0.131 NA 6.974
plot(dx)
 if (FALSE) { # \dontrun{
# Static diagnostics with additional network statistics
dx1 <- netdx(est,
nsims = 1e4, dynamic = FALSE,
nwstats.formula = ~ edges + meandeg + concurrent
)
dx1
plot(dx1, method = "b", stats = c("edges", "concurrent"))
# Dynamic diagnostics on the STERGM approximation
dx2 <- netdx(est,
nsims = 5, nsteps = 500,
nwstats.formula = ~ edges + meandeg + concurrent,
set.control.ergm = control.simulate.formula(MCMC.burnin = 1e6)
)
dx2
plot(dx2, stats = c("edges", "meandeg"), plots.joined = FALSE)
plot(dx2, type = "duration")
plot(dx2, type = "dissolution", qnts.col = "orange2")
plot(dx2, type = "dissolution", method = "b", col = "bisque")
# Dynamic diagnostics on a more complex model
nw <- network_initialize(n = 1000)
nw <- set_vertex_attribute(nw, "neighborhood", rep(1:10, 100))
formation <- ~edges + nodematch("neighborhood", diff = TRUE)
target.stats <- c(800, 45, 81, 24, 16, 32, 19, 42, 21, 24, 31)
coef.diss <- dissolution_coefs(dissolution = ~offset(edges) +
offset(nodematch("neighborhood", diff = TRUE)),
duration = c(52, 58, 61, 55, 81, 62, 52, 64, 52, 68, 58))
est2 <- netest(nw, formation, target.stats, coef.diss, verbose = FALSE)
dx3 <- netdx(est2, nsims = 5, nsteps = 100)
print(dx3)
plot(dx3)
plot(dx3, type = "duration", plots.joined = TRUE, qnts = 0.2, legend = TRUE)
plot(dx3, type = "dissolution", mean.smooth = FALSE, mean.col = "red")
} # }
if (FALSE) { # \dontrun{
# Static diagnostics with additional network statistics
dx1 <- netdx(est,
nsims = 1e4, dynamic = FALSE,
nwstats.formula = ~ edges + meandeg + concurrent
)
dx1
plot(dx1, method = "b", stats = c("edges", "concurrent"))
# Dynamic diagnostics on the STERGM approximation
dx2 <- netdx(est,
nsims = 5, nsteps = 500,
nwstats.formula = ~ edges + meandeg + concurrent,
set.control.ergm = control.simulate.formula(MCMC.burnin = 1e6)
)
dx2
plot(dx2, stats = c("edges", "meandeg"), plots.joined = FALSE)
plot(dx2, type = "duration")
plot(dx2, type = "dissolution", qnts.col = "orange2")
plot(dx2, type = "dissolution", method = "b", col = "bisque")
# Dynamic diagnostics on a more complex model
nw <- network_initialize(n = 1000)
nw <- set_vertex_attribute(nw, "neighborhood", rep(1:10, 100))
formation <- ~edges + nodematch("neighborhood", diff = TRUE)
target.stats <- c(800, 45, 81, 24, 16, 32, 19, 42, 21, 24, 31)
coef.diss <- dissolution_coefs(dissolution = ~offset(edges) +
offset(nodematch("neighborhood", diff = TRUE)),
duration = c(52, 58, 61, 55, 81, 62, 52, 64, 52, 68, 58))
est2 <- netest(nw, formation, target.stats, coef.diss, verbose = FALSE)
dx3 <- netdx(est2, nsims = 5, nsteps = 100)
print(dx3)
plot(dx3)
plot(dx3, type = "duration", plots.joined = TRUE, qnts = 0.2, legend = TRUE)
plot(dx3, type = "dissolution", mean.smooth = FALSE, mean.col = "red")
} # }